High prevalence of urinary schistosomiasis in a desert population: results from an exploratory study around the Ounianga lakes in Chad - Infectious Diseases of Poverty - Infectious Diseases of Poverty - BioMed Central
Study site and study population
The study was carried out in January 2019 around the lakes of Ounianga and in the two settlements of Ounianga Kebir and Ounianga Serir, Ennedi Ouest province in Northern Chad (Fig. 1). The lakes lie around 40 km apart from each other within the hyper-arid Sahara desert with high daytime temperatures and less than 5 mm of rainfall per year. The lakes are fed by an underground aquifer, thereby maintaining the fresh water in some of the lakes despite the enormously high evaporation rates. Therewith, the lakes of Ounianga represent a unique hydrological system [18].
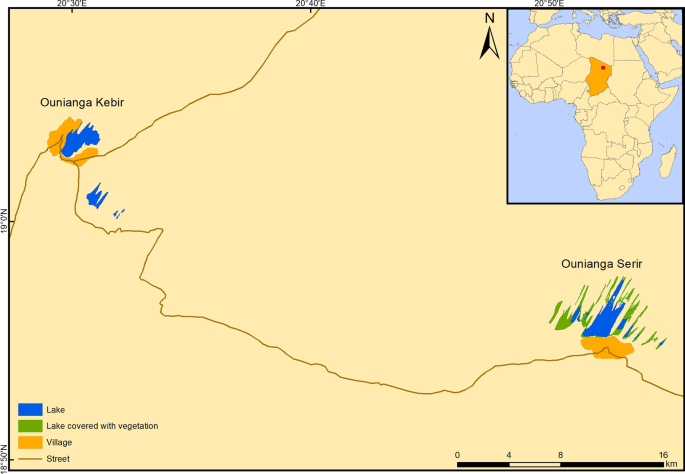
A map showing the lakes and the settlements of Ounianga Kebir and Ounianga Serir in Northern Chad
The official population estimates according to the latest national population census for Ounianga Kebir stand at around 9000 people and for Ounianga Serir at about 1000 people [19]. The majority of these people belong to one of the three predominant ethnolinguistic groups of the Tedega, Dazaga Toubou and the Zaghawa. Their main occupational activities include pastoralism, natron mining and trade. In both communities, the primary schools are operational, yet not the secondary schools. The only functional health centre of the Ounianga district is located in Ounianga Kebir and its catchment population is estimated to include 30,000 people. Ounianga Serir has no functional health centre; the population has set up a health post to provide basic health services to the community members.
Parasitological survey
The resident population of Ounianga Kebir and Ounianga Serir, older than 5 years of age, were eligible for participation. The sample size was calculated using Epi Info 7.1.3.3 (Centre for Disease Control (CDC), Atlanta, GA 30333, USA). Parameters used were "population survey" with two-sided confidence intervals of 95%, an expected frequency of 50% and a population size of 10,000, resulting in a sample size of 370. Proportional to the total population estimates, the targeted sample size repartition was 330 people in Ounianga Kebir and 40 people in Ounianga Serir. At household level and at the primary schools, individuals were randomly selected by applying the spatial sampling method from the Expanded Programme on Immunization (EPI) of the WHO as previously published [20]. After obtaining oral consent from each selected individual, or in case of children from their caretakers, they were asked to produce a urine sample around noon, when S. haematobium egg excretion is known to peak [21]. A mobile field laboratory was set up at the health centre, and health post, respectively. The urine samples were analysed for haematuria by reagent strip testing (Hemastix; Siemens Healthcare Diagnostics GmbH; Eschborn, Germany) and classified as negative, light and severe haematuria as outlined by the testing handbook. Subsequently, 10 ml samples were subjected to urine filtration using a syringe and pressed through a 13-mm diameter filter holder containing a 20-μm wire-mesh filter (Sefar AG; Heiden, Switzerland), followed by microscopic screening of the filter content for the presence of S. haematobium eggs. The infection intensity was determined according to the 3 categories defined by the WHO as no infection (0 egg/10 ml urine), mild infection (1–49 eggs/10 ml urine) and heavy infection (50 or more eggs/10 ml urine) [22]. A point-of-care circulating cathodic antigen (POC-CCA) urine cassette test (Rapid Medical Diagnostics; Pretoria, South Africa) was performed to screen for S. mansoni infections. The POC-CCA test applies a lateral flow principle and allows to detect adult Schistosoma worm CCA in the hosts' urine by adding a colloid carbon conjugate of a monoclonal antibody to the sample, and has been validated before for its performance under extreme environmental conditions as those occurring in the Sahara [23, 24].
Qualitative survey
In both communities, one focus group discussion (FGDs) with men and one with women were organized. Additionally, one FGD was organized with the staff of the health centre in Ounianga Kebir. In Ounianga Serir, an in-depth interview (IDI) was carried out with the person responsible for the health post. The topics covered by the interview guides were disease priorities and priority health issues, perceptions and health seeking behaviour. Both FGDs and IDIs were assisted by an interpreter who translated the conversation from Arabic to French, allowing the study team to take notes. Digital recordings of the FGDs and IDI were transcribed and translated into French, integrating the notes taken during the FGDs or IDIs.
Malacological survey
At both settlements, individual community members and school-aged children were asked to guide the team to all frequented human-water contact sites. At each site, GPS coordinates and the water parameters temperature (°C), pH, conductivity (µs/cm) and dissolved oxygen (mg/L) were recorded, using a portable multimeter (Hach®, HQ40D, Loveland, USA). For turbidity, a turbidimeter was used [Formazin Nephelometric Units (FNU); Hach®, 2100P Iso]. The snail sampling was performed adhering to standard protocols. In short, for 15 min, all aquatic snails were collected by one person using a scoop or forceps to detach them from aquatic and subaquatic plants [25]. Subsequently, the snails were placed on wet cotton in petri dishes, and transferred to the field laboratory. Snails were identified to the genus or, if possible to species level on site. At midday, each collected snail identified as intermediate host species was placed in a well plate filled with bottled drinking water and exposed to daylight for three hours to induce cercarial shedding [26]. The snail size (in mm) and weight (in mg) was measured using a calibre and balance, respectively. Thereafter, all snail specimens were conserved in 70% ethanol, and shipped to the National History Museum, London (NHM) for molecular analysis.
Snail species confirmation using molecular methods
The snail samples selected for the molecular analyses represented individuals from each collection site. On arrival at the NHM, the snail species identification was confirmed based on morphological characters and samples re-spirited (absolute ethanol) for incorporation into the Schistosomiasis Collection at the Natural History Museum (SCAN) [27]. Photographic images were taken of the snail shells prior to DNA extraction. Specimens were placed in TE buffer (10 mmol/L Tris, 0.1 mmol/L EDTA) pH 7.4 for one hour in order to remove any remaining ethanol from within the tissue, which might interfere with subsequent extraction steps [28]. Total genomic DNA was isolated from head/foot tissue using the DNeasy Blood and Tissue kit (Qiagen, UK) according to manufacturer's instructions. DNA was eluted into 200 μl sterile water.
Amplification of Cox1 fragments of snail DNA
A polymerase chain reaction (PCR) amplification of a partial cytochrome oxidase 1 (Cox1) sequence was performed using primers LCO1490 (5′-GGTCAACAAATCATAAAG ATATTGG-3′ forward) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ reverse) [29]. PCR investigations and sequencing conditions were chosen as previously outlined [28, 30].
Checking of sequence data
The electropherograms produced were checked and Cox1 sequences edited using Geneious, version 11.0.5 (http://www.geneious.com) [31]. Sequences were compared to database entries by performing BLAST searches via the National Center for Biotechnology Information against GenBank and EMBL sequence databases; and aligned with reference material.
Sequencing of Schistosoma spp. eggs in urine
Positive urine samples from Ouinanga Kebir were combined into seven different pools of 8–12 ml respectively one pooled sample of 12 ml for the villages of Ouinanga Serir. Samples were shipped to the diagnostic centre of the Swiss Tropical and Public Health Institute (Swiss TPH) in Basel, Switzerland for further processing. There, each pool was centrifuged at 3000 × g for 10 min. Exactly 500 µl of the pellet was re-suspended and transferred to a 2 ml tube containing garnet beads. After addition of 1 ml PBS, the sample was centrifuged 1 min at 13,000 × g and the supernatant was discarded. The pellet with the garnet beads was frozen 30 min at −80 °C and further processed as described by Barda and colleagues [32, 33]. Samples were first tested by simplex generic Schistosoma spp. 28S real-time PCR amplifying S. mansoni, S. haematobium, S. intercalatum, S. bovis [34] and additionally S. japonicum because of modifications added to the second reverse primer of the assay (Additional file 1: Table S1). The reaction mix contained 1 × TaqMan GenExpression MasterMix (ThermoFisher Scientific, Basel, Switzerland), 800 nmol of forward primer, 400 nmol of each reverse primer and 200 nmol of probe. The samples were subsequently tested by a duplex real-time PCR for the presence of a specific S. mansoni TRE region and of S. haematobium dra1 sequence [34, 35]. Each reaction mix contained 1 × TaqMan GenExpression MasterMix (ThermoFisher Scientific, Basel, Switzerland), 800 nmol of each primer, 200 nmol each probe (Additional file 1: Table S1). The thermoprofile of all assays on the QuantStudio5 (ThermoFisher) consisted of 2 min at 50 °C, 10 min at 95 °C followed by 45 cycles of 15 s at 95 °C and 1 min at 58 °C. The specificity of all assays was previously tested on a variety of DNA from stool and blood samples including: Ascaris lumbricoides, Blastocystis hominis, Cryptosporidium spp., Dientamoeba fragilis, Encephalitozoon spp., Endolimax nana, Entamoeba coli, E. dispar, E. histolytica, E. moshkovskii, E. polecki, Enterocytozoon bieneusi, Giardia lamblia, Hymenolepis nana, Iodamoeba bütschlii, Sarcocystis spp., Taenia spp., Strongyloides stercoralis, Trichuris trichiura, Plasmodium falciparum, P. vivax, P. malariae, P. ovale, Trypanosoma cruzi, T. brucei, Leishmania spp. and was found to be 100% specific. Analytical limit of detection (LOD) was tested by a plasmid dilution row ranging from 107 to 10–1 plasmids/µl containing an insert with the sequence of the Schistosoma real-time PCR product, and was found to be at 10 plasmids/µl for all assays. On each real-time PCR plate and for each target we included negative and positive low-copy plasmid controls.
Subsequently, all samples were tested by classic PCR of the COX gene of S. haematobium and S. bovis as modified from Boon and co-workers (Additional file 1: Table S1) [36]. The reaction mix contained 1 × HotStarTaq Plus Master Mix (Qiagen, Hilden, Germany), 800 nmol of each primer, 5 µl DNA in a total reaction volume of 50 µl. The thermoprofile consisted of 5 min at 94 °C followed by 40 cycles of 40 s at 94 °C, 40 s at 58 °C and 1 min at 72 °C and a final step of 10 min at 72 °C. After visualization on a 2% agarose-gel, the positive sample of the S. bovis-COX PCR was sent for Sanger sequencing with the primers of amplification at Microsynth AG (Baldach, Switzerland). The sequence was then compared to database entries by performing BLAST searches via the National Center for Biotechnology Information. The sequence is accessible in GenBank under the number: MW937895. A table listing primers and probes is accessible in the supplementary information (Additional file 1: Table S1).
Statistical analysis
Descriptive statistics of parasitological and malacological data was performed using STATA version 16.0 (STATA Corp Inc., TX, USA) and ArcGIS (Version 10.7.1.; ESRI Inc. ArcMap™ 10.7, Redlands, CA, USA). Qualitative data analysis included the full review of all transcripts, followed by a descriptive and explorative thematic analysis.
Comments
Post a Comment