Emerging infectious disease surveillance using a hierarchical diagnosis model and the Knox algorithm | Scientific ... - Nature.com
Abstract
Emerging infectious diseases are a critical public health challenge in the twenty-first century. The recent proliferation of such diseases has raised major social and economic concerns. Therefore, early detection of emerging infectious diseases is essential. Subjects from five medical institutions in Beijing, China, which met the spatial-specific requirements, were analyzed. A quality control process was used to select 37,422 medical records of infectious diseases and 56,133 cases of non-infectious diseases. An emerging infectious disease detection model (EIDDM), a two-layer model that divides the problem into two sub-problems, i.e., whether a case is an infectious disease, and if so, whether it is a known infectious disease, was proposed. The first layer model adopts the binary classification model TextCNN-Attention. The second layer is a multi-classification model of LightGBM based on the one-vs-rest strategy. Based on the experimental results, a threshold of 0.5 is selected. The model results were compared with those of other models such as XGBoost and Random Forest using the following evaluation indicators: accuracy, sensitivity, specificity, positive predictive value, and negative predictive value. The prediction performance of the first-layer TextCNN is better than that of other comparison models. Its average specificity for non-infectious diseases is 97.57%, with an average negative predictive value of 82.63%, indicating a low risk of misdiagnosing non-infectious diseases as infectious (i.e., a low false positive rate). Its average positive predictive value for eight selected infectious diseases is 95.07%, demonstrating the model's ability to avoid misdiagnoses. The overall average accuracy of the model is 86.11%. The average prediction accuracy of the second-layer LightGBM model for emerging infectious diseases reaches 90.44%. Furthermore, the response time of a single online reasoning using the LightGBM model is approximately 27 ms, which makes it suitable for analyzing clinical records in real time. Using the Knox method, we found that all the infectious diseases were within 2000 m in our case, and a clustering feature of spatiotemporal interactions (P < 0.05) was observed as well. Performance testing and model comparison results indicated that the EIDDM is fast and accurate and can be used to monitor the onset/outbreak of emerging infectious diseases in real-world hospitals.
Similar content being viewed by others
A novel data-driven methodology for influenza outbreak detection and prediction

Thirty-day hospital readmission prediction model based on common data model with weather and air quality data
Forecasting influenza-like illness trends in Cameroon using Google Search Data
Introduction
Emerging infectious diseases are defined as "new, emerging, or drug-resistant infectious diseases, the occurrence of which in the population has increased in the past 20 years or for which there are indications that their incidence may increase in the future"1. Emerging infectious diseases mainly include new diseases, existing diseases emerging in a new area or population, reintroduced old diseases, previously clinically mild diseases increasing in severity, and previously preventable or treatable diseases becoming uncontrolled or treatment-resistant2. Emerging pathogens include bacteria, viruses, parasites, chlamydia, rickettsia, spirochetes, and mycoplasma, among which viruses cause the largest number of emerging infectious disease cases. Emerging infectious diseases, which can cause serious regional or international public health concerns, are one of the most critical public health challenges faced by humankind in the twenty-first century. In the past 30 years, at least 40 emerging infectious diseases have been detected worldwide1, and their number is rapidly increasing, posing a considerable threat to human life and health. For example, SARS raged in 2002–2004, the Ebola virus broke out in Africa in 2013–2016, Zika virus cases were detected in 2016, yellow fever was detected in 2016–2018, and so on. Emerging infectious diseases possess the inherent infectivity and prevalence of infectious diseases, they have complex origins, and tracing the sources of these diseases is arduous. Furthermore, they tend to be widespread and difficult to control, making them uniquely uncertain and unpredictable. Owing to these characteristics, these diseases cause extensive harm, as there are no existing guidelines regarding their prevention and control. For example, the coronavirus disease (COVID-19) that occurred in 2019 transmitted rapidly and spread widely, turning into a pandemic, thereby damaging societal and economic stability worldwide3. Accordingly, monitoring and early warning of emerging infectious diseases are critical steps towards preventing their spread. Forecasting of emerging infectious diseases and generating an early warning system to avoid their outbreaks are major challenges.
At present, most emerging infectious diseases are discovered because of abnormalities noticed by clinicians. After observation, laboratory biological testing, clinical treatment, and so on, certain disease symptoms are identified and categorized as symptoms of emerging infectious diseases. For example, COVID-19 was discovered when clinicians, who diagnosed the symptoms, reported that many patients were employees of the Huanan Seafood Wholesale Market, and bioinformatics testing showed that the pathogen was a new type of coronavirus4. Thus, clinical symptom monitoring and access to the complete medical information of a patient can aid in identifying emerging infectious diseases. Medical institutions are the first line of defense for diagnosing and treating such diseases. Unlike public health monitoring to identify known infectious diseases, timely monitoring of abnormal symptoms and phenomena by medical institutions during infectious disease outbreaks is more beneficial for emerging infectious disease management5. Continuous improvements in hospital medical information systems over the years have resulted in the accumulation of a large amount of medical data, including health data, patient information, medical records, inspection records, imaging records, and cost information. In a medical institution with an average daily outpatient volume of 15,000, the medical data volume increases by 50 Gb daily6. In general hospitals in China, daily outpatient visits generally exceed 10,000. Therefore, in addition to seeing patients, doctors in medical institutions are expected to identify clues of emerging infectious diseases from many complex and related medical records, and this task is extremely difficult. Furthermore, clinicians need to follow a specific set of rules and regulations while identifying infectious disease risks, performing active identification and legal reporting, and it is objectively difficult for clinicians to take the initiative to provide early warning in the absence of relevant background information. To address these challenges, a big-data-based effective prediction model that can extract features from a large volume of data, perform data mining, and conduct simultaneous time and space monitoring is urgently required.
In general, medical institutions predict the prevalence and outbreak of infectious diseases. Unlike traditional statistical methods, machine learning and deep learning are data driven. To date, several studies on disease diagnosis, hospitalization, prediction of treatment duration, etc. have been conducted using health data obtained from medical institutions7,8. These reported studies have revealed several significant theoretical and practical results, which have propelled further research in this field. In recent years, early warning generation and prediction of the onset of infectious diseases have come under the spotlight because of the advances in big data and machine learning; for instance, Lee et al., Feng and Jin, and Wang et al. 9,10,11 predicted the onset trend of known infectious diseases. Based on the details, it can be concluded that medical institutions acquire health data during the early stages of the disease as well as are the first points-of-contact for patients infected with various known/unknown infectious diseases. In addition, they have the basic conditions for analyzing the outbreak of emerging infectious diseases. Thus, a combination of machine learning and deep learning will facilitate medical institutions in early detection of the emerging variants of infectious diseases, thereby enabling an effective and timely containment to prevent their outbreak. Thus, this study was designed to construct an emerging infectious disease identification framework based on the real and complete medical records of hospitals, with hierarchical diagnosis model (EIDDM) and the Knox method for spatial cluster as the cores, using machine learning. Considering the accuracy and computational efficiency of data collection in actual medical institutions, a hierarchical diagnosis model, namely the emerging infectious disease detection model (EIDDM), which combines the TextCNN-Attention and LightGBM algorithms, was developed. This proposed framework is suitable for spatial–temporal monitoring of infectious diseases in medical institutions and is anticipated to mitigate the existing key issues related to emerging infectious disease management. The primary objectives and contributions of the present study are as follows:
Analysis of the current research methods on the identification of emerging infectious diseases, and summary of the currently known key issues, including the reported data of symptom events, inadequate reporting of active behavior, and disconnection between non-clinical data (such as online search data) and real medical cases.
Based on the analysis of datasets acquired from multiple medical institutions, structured and unstructured data are applied to various models for separate processing, and a special processing is carried out for different types of features to utilize all the features completely. These processes provide separate datasets for training as well as real-world application of the developed model.
The proposed EIDDM considers both model recognition and online reasoning efficiency. The average emerging-infectious-disease prediction accuracies of the first- and second layer models were 90.47% and 86.93%, respectively. The prediction time of the online reasoning for a single medical record is only 27 ms, which is remarkably less than that required for one-hot encoding of a single medical record in previous studies (68 ms) and is more suitable for real-time scenarios in real clinical medical institutions.
Cluster analysis was performed using the Knox method, which does not require population migration and total population data. A cluster analysis is an effective supplement to EIDDM, which is a hierarchical diagnosis model, to determine the existence of a spatial cluster after discovering the emerging infectious disease cases. This step reduces the misjudgment rate of individual case samples when the model is applied to the data obtained from real medical institutions.
The rest of the paper is organized as follows: Section "Related work" presents and describes the previous studies reported in this field. Section "Materials and methods" discusses the data processing and modeling approaches employed in this study. Section "Results" elucidates the experimental results and compares them with other state-of-the-art research strengths and limitations. Section "Discussions" discusses the rationale for using the one-vs-rest (OvR) strategy to identify emerging infectious diseases. Section "Conclusion" highlights the major conclusions drawn from the findings of this study.
Related work
Currently, monitoring of emerging infectious diseases is one of the key tasks of public health emergency detection and is accomplished mainly through the collection, analysis, identification, and intervention of the occurrence, spread, and source of infectious diseases in the population. According to Christaki et al. 12, infectious disease monitoring is divided into event-based surveillance, web-based real-time surveillance, social media monitoring, and new technologies in pathogen discovery. Event-based surveillance is primarily organized by health authorities. In China, infectious diseases are divided into categories A, B, and C, and doctors in medical institutions are required to report through the national infectious disease reporting system within a limited time 13. This system enables medical institutions to act as monitoring sentinels to report and review patients with infectious or suspected infectious diseases. The system also aids health departments and disease control systems in rapidly analyzing and judging any epidemic situation. The observations made in the initial stage of the novel coronavirus epidemic revealed that the system is only limited to daily monitoring and reporting of known infectious diseases. These attributes cannot meet the requirements of information acquisition, early warning, and disposal of emerging infectious diseases 14. Furthermore, the elements involved in the surveillance of emerging infectious diseases are diverse and complex, and the fixed indicator combination of traditional disease surveillance is not necessarily applicable to these emerging infectious diseases. Previously, some researchers 15,16 conducted surveillance of influenza, influenza-like illnesses, and severe respiratory illnesses based on the symptom surveillance system, which caused a large analysis bias due to the inevitable gap between the symptoms and the actual diagnosis. However, only considering symptoms without incorporating the time and space information cannot satisfy the prediction accuracy of regional infectious disease incidences.
In online-data-based surveillance, the data are chiefly obtained from non-medical institutions in the form of online search engine data, news data, or social data to monitor diseases with potential epidemic risks as well as infectious diseases with seasonal activities. Ref. 17,18 report the use of results obtained from web search engines as the data sources for infectious disease symptom monitoring. In these two reported studies, correlation analysis was conducted between the frequency of the related search terms and the actual number of people with symptoms to track the spread of regional infectious diseases. According to Ref. 19, Google's prediction of flu trends can be 1 to 2 weeks ahead of that of the Center for Disease Control and Prevention. Juhyeon et al. 20 used Medisys to collect internet articles related to infectious diseases to predict the outbreak of infectious diseases using a support vector machine-based model. However, because search engines and news methods require many search queries, in terms of disease coverage, they only support conventional symptoms that the public can describe, such as fever, vomiting, diarrhea, etc., and cannot monitor abnormalities in inspections. In addition, there is a disconnect between the online data and real medical records of medical institutions, and thus, the online method cannot be truly applied in the monitoring scenarios of medical institutions. From the perspective of spatiotemporal monitoring of infectious diseases, online data monitoring facilitates a general large-scale prediction across the country, which cannot be analyzed from the perspective of a spatial cluster. However, a spatial cluster can be located by analyzing the home and work addresses provided in medical records.
With the development of machine learning and neural networks, several methods for early warning and prediction of infectious diseases have been reported to date. Wilkinson et al. 21 adopted a statistical process control method, because the data should be independent, and the known parameters should follow a normal distribution. However, the actual transmission mechanism of infectious diseases cannot be completely independent; a certain correlation between the cases is expected. Nevertheless, the machine-learning-based prediction methods exhibit some shortcomings. For instance, although a decision tree can be easily interpreted, a single tree is more sensitive to noise data and has a poor generalization ability. However, an integrated model based on the decision tree can overcome the shortcomings of a single tree. Further, the Bayesian method has a simple logic, is easy to implement, and performs well when the features in the correlation are relatively small. However, the algorithm involves independent assumptions of the feature conditions, and performs poorly when there are many features, and the correlation between the features is large22.
Presently, only a few studies on early warning of emerging infectious diseases are available. Li et al. 23 extracted the features from historical medical records of various known diseases and constructed a disease probability map. When the probability of a new patient, becoming infected by each known disease type, is less than the threshold set by each known disease, the case of emerging disease types is evident. Although this method considers different diseases independently, it ignores the complexity of the diseases in real medical scenarios. Currently, thousands of common diseases are known, and the relationships among them are complex. Thus, even if the training covers various known diseases, the complex relationship among them has not yet been effectively accounted for by the existing machine-learning-based prediction models. In our previous study 24, we used 20,620 real infectious disease datapoints obtained from a large hospital from 2012 to 2022, including outpatient and inpatient sample data, to construct a multi-infectious disease diagnosis model (MIDDM) and obtained 740,000-dimensional feature data, and then performed model training after a sparse data densification processing. In addition, a residual network and an attention mechanism were introduced into the MIDDM to improve the model performance. However, due to the large feature dimension of this method (obtained after the one-hot encoding), the dense expression vector needs to be calculated first through the dense network and finally judged by the classification model. Thus, numerous model parameters are involved in this complicated process. Further, the prediction time has a remarkable impact. The prediction time for a single medical record reaches 68 ms, and the average response time is higher than 400 ms when the number of concurrencies is 100.
Therefore, to realize an effective early warning strategy for emerging infectious diseases via data monitoring, it is necessary to develop an automated monitoring approach that overcomes the issues related to manual monitoring, which involves event and statistical analyses and solely relies on manual reporting of symptoms. In this study, we used a hierarchical model to assess the probability of a single sample being an emerging infectious disease case, and combined the proposed model with the Knox method to analyze clusters. The method considers the case attributes as well as rapidly determines the probability of occurrence of infectious diseases. Further, to achieve data source tracking with high fidelity, it is necessary to utilize the complete existing data of hospitals in combination with real cases for monitoring. In this study, all the medical records obtained from multiple medical institutions were used, and the data time information was retained through time sequencing. To solve the problem of low performance caused by dimension explosion after one-hot coding in the early stage, we adopted the word vector model, word2vec, for data pretraining. Combined with the LightGBM model, word2vec can directly read the word vector in the subsequent model training without the need to calculate the dense vector expression of the current input to improve the prediction efficiency.
Materials and methods
This section describes the selection of datasets, data preprocessing, model architecture, classification methods, performance evaluation methods, experimental tools and setup, and validation methods. Figure 1 depicts the artificial intelligence process proposed here for the surveillance of emerging infectious diseases.
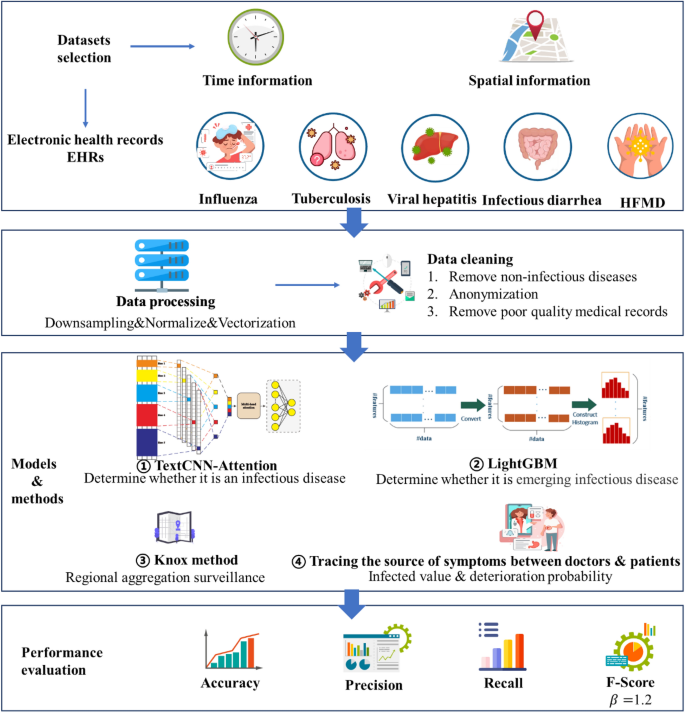
Proposed approach.
Dataset selection
The data were obtained from five medical institutions in Beijing, China: three in the Haidian District (Headquarters hospital, North hospital, and Party School hospital), one in the Shunyi District, and one in the Daxing District. The location distribution meets the spatial heterogeneityspatial heterogeneity requirement 25. 'Spatial-specific' refers to characteristics that distinguish things or phenomena in each spatial location from those in other locations. The medical data of five institutions, including all outpatient, emergency, and inpatient data obtained from January 1, 2012 to December 31, 2021 related to the 59 infectious diseases 26 highlighted by the China Center for Disease Control and Prevention, are considered. By matching the diagnosis name with the ICD-10 code, all the diseases and sub-diseases belonging to the infectious disease category were included in the dataset, and senior medical experts removed some non-infectious sub-diseases such as thyroid tuberculosis and renal tuberculosis. In order to obtain high-quality training data, names such as diseases and symptoms were standardized and the associated data were verified and subjected to an integrity check. Finally, 37,422 and 9,325,680 cases of infectious and non-infectious diseases, respectively, were identified as listed in Table 1. Some infectious diseases, such as pestis, did not occur, and hence, they are not listed in this table.
Owing to the large difference in the number of infectious and non-infectious diseases, stratified sampling was conducted. Most infectious diseases belong to obvious categories (e.g., most of them are respiratory, skin, or digestive diseases), while a few of them are related to orthopedics. Therefore, stratified sampling was carried out based on the departments from which the samples were obtained. For example, if pulmonary tuberculosis belongs to the respiratory department, then the other diseases identified in the data obtained from the respiratory department, such as pneumonia, lung cancer, chronic obstructive pulmonary disease, and bronchiectasis, are randomly selected. In addition to restricting the selection of medical records on non-infectious diseases obtained from the departments where the infectious diseases are evident, we also conducted random down-sampling of the non-infectious diseases. This step enabled the inclusion of a relatively small number of medical records on non-infectious diseases, and the number of non-infectious-disease medical records used as the training samples was 1.5 times that of the infectious cases. After the stratified and random down-sampling, 56,133 cases of non-infectious diseases were finally included. The screening process of the patients' medical records is illustrated in Fig. 2.
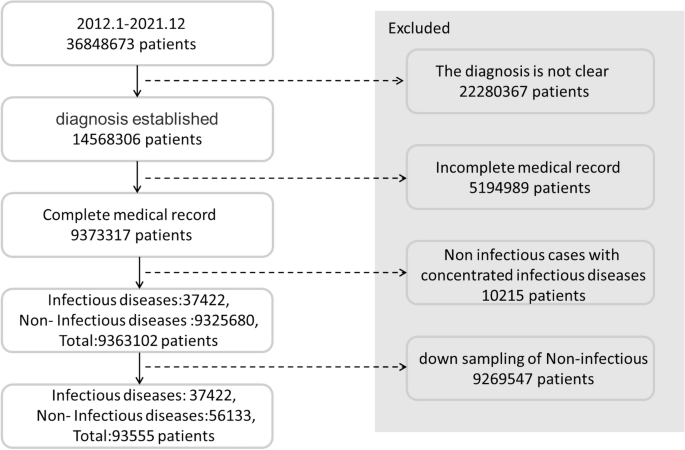
Flowchart of enrollment.
To ensure maximum utilization of the information on important factors related to infectious diseases, the data used in the training model include basic patient information, time information, spatial information, patient medical records, and so on as presented in Table 2. Notably, to prevent information leakage during model training, we eliminated the test items that can clearly indicate the type of infectious disease in the test report, e.g., the new coronavirus nucleic acid test items in the medical records of COVID-19 patients. The diagnosis was used as the label of the sample; the work and home addresses were used as the key information to trace the address of the infectious disease patient; and the remaining extracted fields were used as the input features of the subsequent diagnosis model.
Data preprocessing
Data extraction
Electronic medical records mainly include data in both structured and unstructured (free text) formats. The patient's basic information, diagnosis, outpatient diagnosis, and test report form the structured data of the electronic medical record, while the admission record, outpatient medical record, and examination report become the unstructured data.
Structured data directly extracts the value of the corresponding field from the electronic medical record. Specifically, information such as age, age unit, visit time, home address, and work address are extracted from the front page of the medical record; information such as the patient's diagnosis name and code are extracted from the home page or outpatient diagnosis; inspection time, inspection value, inspection scope, and other information are obtained from the report page. Then, abnormal value processing, number normalization, and standardization are carried out for continuous variables; for example, the abnormal value processing of age will filter out the ones that deviate from the normal age range. Then, according to the different age units (year, month, week, etc.), the age value is uniformly converted into the age unit; each inspection result in the inspection report is classified as "high," "low," or "normal" according to their normal value range. In addition, the extracted discrete variables such as diagnosis name and inspection sub-item name are unified into standard names according to the mapped relationship between the aliases and standard names in the knowledge base. Finally, the processed field data are sorted into a text sequence.
For unstructured data, it is first necessary to extract information such as the chief complaint of the admission record, inspection conclusions, and inspection findings in the inspection report 27,28. In this case, we extract various entities (such as time, symptoms, diseases, signs, drugs, inspections, etc.), and then standardize each entity based on the British Medical Journal's Best Practices 29 knowledge base, and finally obtain a data sequence. Subsequently, all the data sequences extracted from the patient's medical record are spliced according to the order of the document and inspection times to finally form the patient's input sequence for the model. The text merging process is illustrated in Fig. 3.
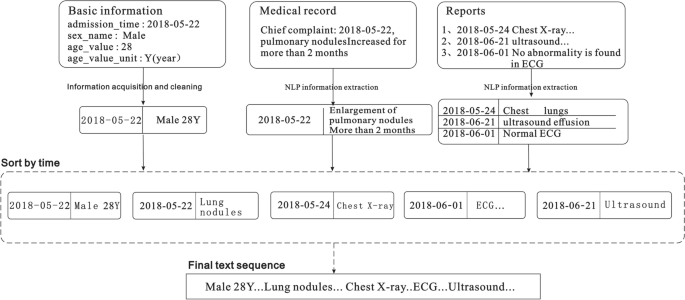
Extraction and serialization of information from electronic medical records.
Data vectorization
To use the above-mentioned serialized data to train the model, it is also necessary to vectorize the sequence text and convert each word in the sequence into a "computable" and "structured" vector before including the data into the model as input. Word2vec, a relatively common tool for training word vectors 30, can transform words into dense, low-dimensional real value vectors, which capture useful syntax and contextual semantics 31. Therefore, we used the Skip gram in word2vec to train the word vector model. Before training the word vector, each word in the input sequence is assigned a unique identifier to identify the word. This identification number is based on the sequence of word frequency in the corpus, and the word is labelled with the highest frequency; for example, the word "male" is assigned a unique identification number < 360 > . In this study, the typical numerical values in the medical records were spliced into a new word by connecting the names, values, and units, and then a unique identifier was assigned to the spliced output; for example, "alkaline phosphatase 102 U/L" is assigned a unique identifier 891. Therefore, the Chinese word sequence is converted into a word index sequence. Then, the word vector model is trained based on the obtained word index sequence. When training the word vector model, we set the word vector dimension to 100, and a window size of 5 is adopted. In addition, considering that the time complexity of the Skip gram is expressed as window size × thesaurus size, a large number of words in the training set increase the computation time. Therefore, we set min_ count = 2 in the model, and ignore the low-frequency words that only appear once or twice.
When vectorizing sequence data, we first calculate the length of all the input sequences and denote the length L of its 98% quantile as the final length of each sequence data. Then, each input sequence is truncated and filled with 0. Next, the unique identifier corresponding to each word in the sequence is converted into a word vector representation according to the trained word vector object. Finally, all the word vectors are spliced together to form an input vector of length L × 100. The process of converting medical records into a vector is depicted in Fig. 4.
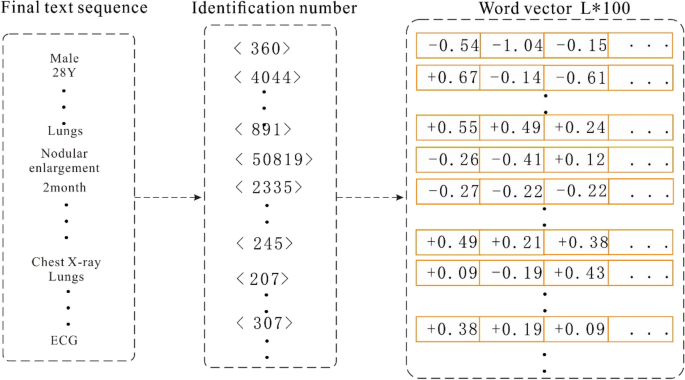
Word vector conversion.
Unbalanced dataset processing
To alleviate the impact of data imbalance on the model results, down sampling was performed in the first layer model in this study. The number of medical records on non-infectious diseases is 1.5 times of that on infectious diseases. In the training of the two-layer model, we added category weight processing to provide categories with fewer samples a higher calculation weight, expressed by formula (1), and use them extensively for model training:
where \({w}_{k}\) represents the weight of the \(k\) class, \({N}_{all}\) represents the total number of samples in the dataset, \(C\) denotes the total number of categories (\(C\) = 8), and \({N}_{k}\) represents the number of samples of the \(k\) class. When the weight is not changed, the weight for each category is represented by the average attention degree (\(1/C\)), and the weight calculation formula satisfies:
Category weight × proportion of the number of category samples in the total dataset = Average attention.
Proposed EIDDM architecture
In this study, we mainly used the EIDDM (TextCNN-Attention + LightGBM), a regional clustering analysis, and symptom association of medical staff to predict the outbreak of emerging infectious diseases. Although no current case of emerging infectious diseases such as SARS, COVID-19, monkeypox, etc., was analyzed in this study, their performance is similar to that of the known infectious diseases; further, their characteristics are similar to those of regional clusters of infectious diseases. Therefore, we used a two-layer model to predict the number of people infected with suspected emerging infectious diseases, and then used their contact addresses to evaluate the possibility of existence of regional clusters. The EIDDM flow is shown in Fig. 5. Finally, through the symptom association analysis of medical staff, we assessed the probability of "human-to-human transmission.". The results of the analysis of interpersonal association transmission 32,33,34 are expected to promote further follow-up epidemiological studies on transmission routes, gene sequencing, and prevention and control programs.
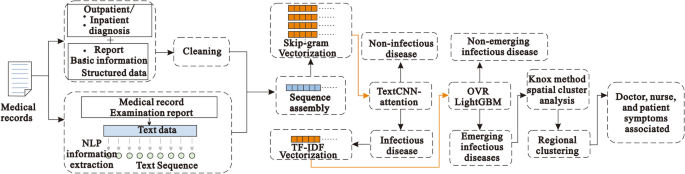
EIDDM flow.
First layer of model with TextCNN-attention
Because of the large amount of medical record data and high-dimensional feature sequence text data in the first layer model used in this study, the model selects the text-based convolutional neural network TextCNN for classification. This network considers both convolution extraction and timing. The word embedding features of the long text sequences are composed of admission records, and thus, the inspection and testing documents have different effects on the classification results. The features that have a greater impact on the model decision are given a greater weight of attention via the Multi-Head Attention mechanism, and then the key and interference features of the text can be distinguished. Therefore, the introduction of the Attention mechanism in TextCNN can effectively enhance the feature extraction capability of the model.
First, zero-padding is used for the filling to ensure that the length of the input feature sequence is consistent in the model. The final maximum sequence length is L (L = 5061), indicating that the input sequence size is 5061 (after the operation: word vector layer × 100). After the word embedding layer, a word embedding matrix with a size of 5061 × 100 is formed. The parameters of the word embedding layer are initialized using the Skip-gram method. Notably, one-dimensional convolution is used in this convolutional neural network. We consider the number of samples, hardware equipment performance, model complexity, case data characteristics, and other factors, and use the grid search 35 method to set multiple values for the same parameter in different value domains and magnitude ranges in descending order. By comparing the accuracy of the trained model using the test set, five filters with dimensions of 1 × 100, 2 × 100, 3 × 100, 4 × 100, and 5 × 100 are set. After the convolution layer, five convolution expressions, L × 100, (L-1) × 100, (L-2) × 100, (L-3) × 100, and (L-4) × 100 are obtained, respectively. Then, we add the max pooling layer to reduce the dimensions of the filter-layer features and splice the pooled vectors; the spliced dimensions are 500. The reduced-dimension spliced pooled vectors are input into the Muti-Head Attention layer with multi-head parameter = 8 and head_dim parameter = 16. Then, we connect a residual (size: 500) with the normalization layer. Finally, the vector is expanded, and in order to prevent overfitting, the dropout loss mechanism is used as the input of the sigmoid layer. The structure of the first layer of the EIDDM is shown in Fig. 6.
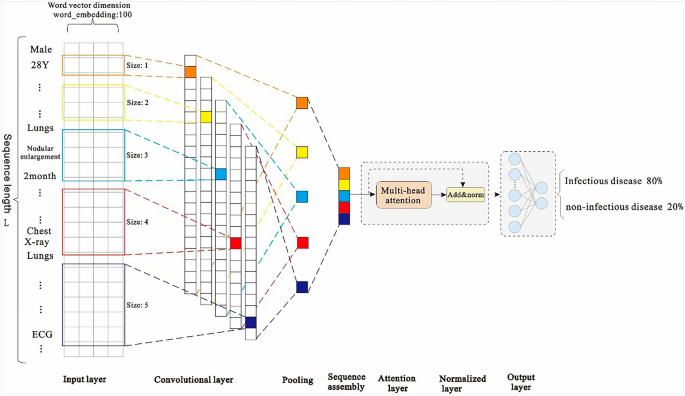
First layer of the ...
Comments
Post a Comment